Distribution of Processing Jobs
Data Visualization
World Wide Web
Data Cataloging
A number of technical obstacles currently impede large scale conservation analyses, including inadequate data management systems, both in terms of design and user interface, for retrieval and manipulation of large volumes of diverse data collected at a variety of space and time scales. Some of our data are original, but many have been acquired from other institutions and individuals. Also, we share many of our data with other parties, increasingly via Internet exchanges. We have encountered a number of problems that will be familiar to others who have built large geographic databases that are accessed by many users with a wide range of expertise. Most importantly, searching for information on a particular theme, time period, or place is laborious; data that we have acquired, as well as those produced locally, are very unevenly documented; often several copies and/or versions of a dataset will coexist on disk and it may be difficult to determine the lineage or processing history of different versions, especially when their creator has left the research team; because our data come from many sources and are distributed widely, maintaining the currency of our database and concurrence with outside users of our data is an enormous challenge. During Year 1 we created a public domain cataloging and browsing tool (PGBIO) based on an existing DBMS (postgres, which we ported to the IBM RISC platforms) and graphical user interface tools (TCL/TK). The tool was extremely effective and useful. A technical paper was written describing the query too and informing researchers how to obtain the software, and distributed to data managers at the U. S. Geological Survey, the World Bank, and the Australian Environmental Resources Information Network (ERIN) who have expressed interest in this cataloging approach.
Our experience with the first version of PGBIO was that it was a very useful product for locating datasets but that it was overly complicated for others to install and operate. The original version relied on postgres, a public domain, object-oriented database management system and TK/TCL as the graphic programming language. For new installations, users had to ftp software from several sites, which were not always stable. Once the software was installed, the system manager had to reconfigure the UNIX server's operating system and maintain a database server not conforming to the SQL database language standard. The executable files for postgres are quite large, and pgbio did not make extensive use of its database management capabilities. Therefore, we redesigned the data cataloging tool without postgres. The second version used simple ASCII flat files for the database. The system utilized the UNIX rcs function to allow simultaneous users to read the catalog while maintaining concurrency and history of the database. Along with the revisions to the software of the cataloging tool, we collaborated with the California Environmental Resources Evaluation System (CERES), a consortium of state environmental agencies, to compile a subset of the items in the federal standards. The new version of our cataloging system incorporates that subset of items to be used as a prototype node of the CERES network.
We then rewrote the data catalog interface in HTML (Figure 25) with PERL CGI scripts to access the database. Since most users are comfortable with a WWW browser interface such as Netscape or Microsoft Internet Explorer, users are comfortable with its look and feel and there is virtually no learning curve to its operation. The new version also provides more display functions to inline display many image format types and text files, while launching helper applications for map data, word processing documents or postscript files. In addition to its familiar feel, an ``autofill'' function has been implemented which automatically fills in all the metadata that can be obtained directly from the dataset. The autofill function eliminates tedious entries for information that is already contained in the dataset itself. This encourages users to take advantage of the powerful tools the data catalog system provides.
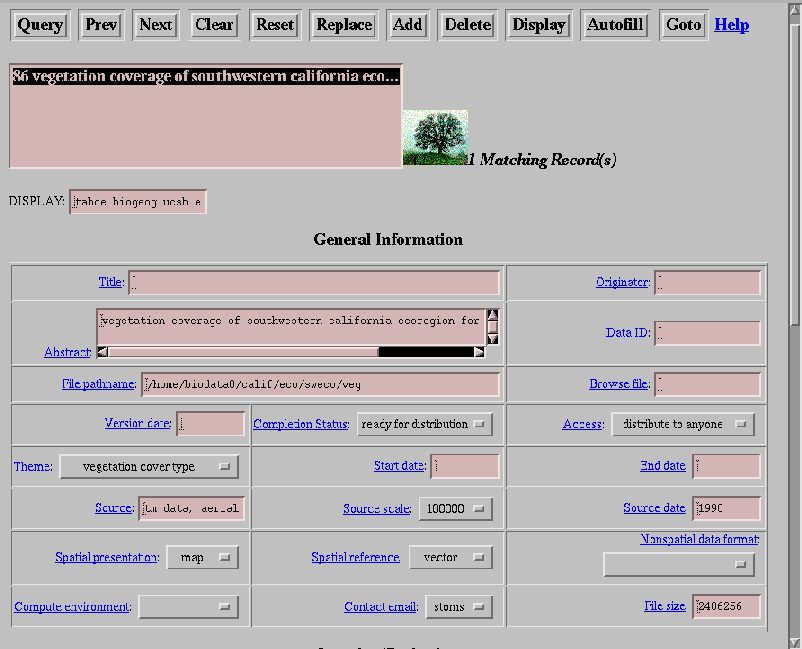
Figure 25. Portion of the HTML version of pgbio interface
Distribution Of Processing Jobs
Some of our GIS and digital image processing tasks require several independent commands to be executed and their results combined. These commands can be easily done sequentially on a single CPU. When there are a lot of commands and/or each command is very CPU intensive, this method can be very time consuming. The user could log in to several machines and execute a few commands on each simultaneously, but this involves more bookkeeping. Also, since some machines are faster than others due to user load of CPU speed, this may be inefficient. The answer has been the development of a job distribution program that remotely executes commands on several computers simultaneously. It is simply a way of automating the process of a user logging in to several machines to execute CPU intensive operations. Virtually any non-interactive UNIX command can be executed on an available server.
The input to the program is a list of UNIX commands that can be contained in a file. When the program is executed it searches for available servers and executes one command in the list on each server simultaneously. When any server finished its command it is sent another until the list of commands has been exhausted. Using this form of job distribution, all the computers can be kept busy and results obtained in a more timely fashion.
Here is one example application that is representative of many GIS and image
processing tasks: A user has 50 maps obtained from an outside data source.
Each map must be imported into a native GIS file format and processed, and
when finished, all maps joined to make a single map. This list of 50 commands
could easily be handled using this form of job distribution. Each command
could be doled out to an idle server so all computers are working on the same
complex task at once. Given the single result is a product of all the steps
involved, this form of job distribution can be loosely coined joint
processing. Using this tool has allowed us to take greater advantage of the
compute power provided by the network of servers and workstations provided by
the IBM-ERP gift.
Data Visualization
Regional biodiversity databases are complex in the number of elements to
understand. The Gap Analysis of California database has many map layers, each
with a large number of attributes. The vegetation database, for instance, has
detailed information on dominant canopy plant species, their relative
abundance, the association with other species as recurring communities, and
canopy closure. Our results with the database suggest that the landscape
database approach will yield useful information for regional conservation
planning that goes beyond that provided by simple vegetation maps. The
database contains more extensive attribute detail than these simpler
land-cover type maps. Much of the information is encoded as alphanumeric
codes related by lookup table to the botanical names or other more readily
understandable description. For instance, to ask what is the distribution of
a plant species that is dominant in the canopy in the Sierra Nevada, the user
would need to know the 5-digit code number of that species and that it could
occur in potentially ten different columns or items in the database.
Unfortunately, this richness of the database makes it more difficult for
novice users to answer their conservation or biogeographical questions. One
of the project objectives was to develop new software to facilitate analysis
of the species data by other botanists, ecologists and biogeographers, and the
general public.
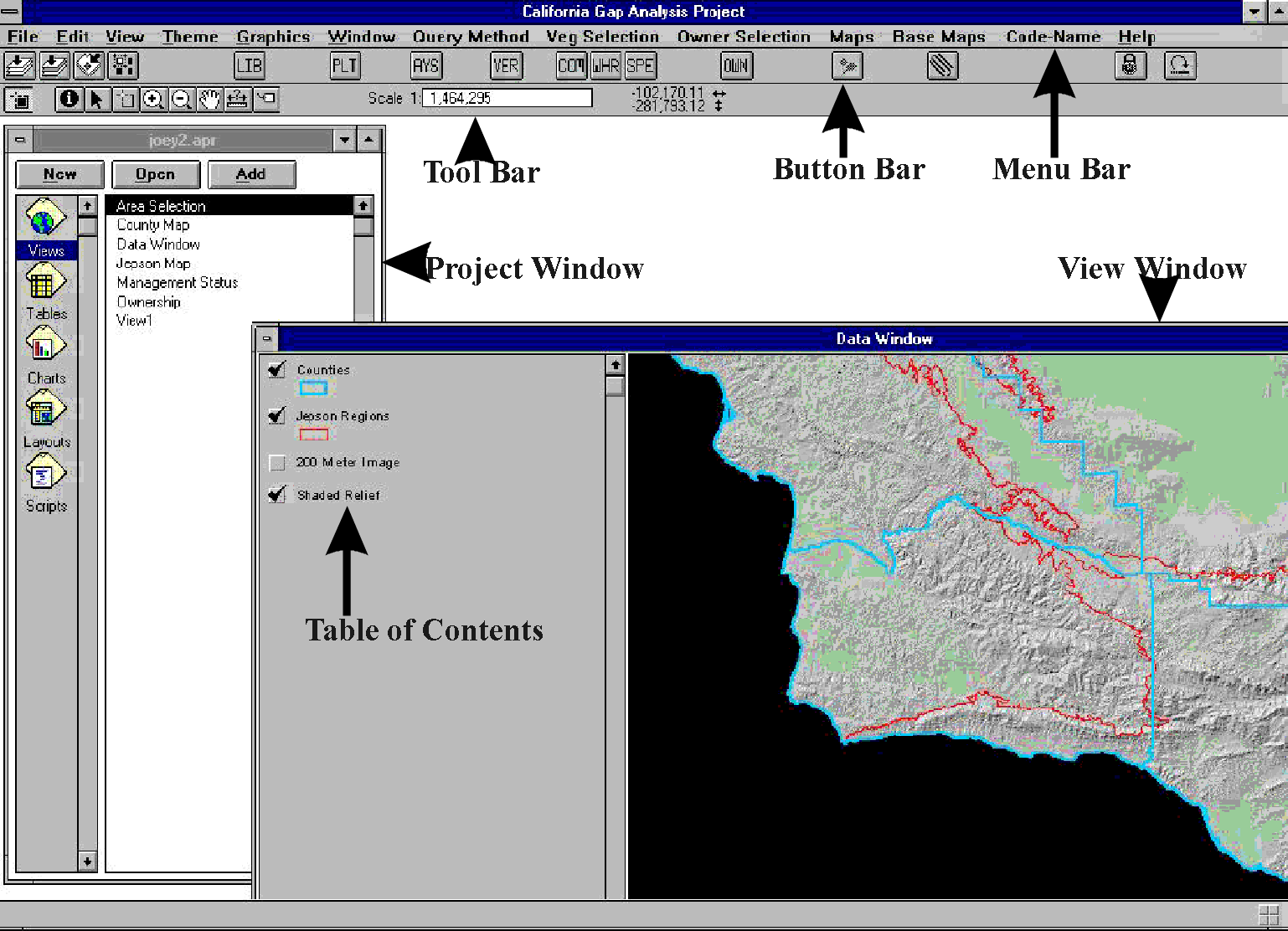
Alternative software tools were explored for developing a better interface to
the GAP database to assist new users in querying the complicated spatial
database. Whereas we originally expected to have to program this interface
from scratch, GIS technology advanced to the point where we could adapt
existing software to the task. Thus we have customized an interface (Figure
26) in ARCVIEW, a commercial desktop map analysis package, using its own
programming module called AVENUE. This interface was designed to interpret
the most frequently posed queries of the database while freeing the user from
having to know the structure of the database or the internal coding of
attributes. The interface will allow the user merely to specify a species and
the interface program will interpret the request into the appropriate database
query. Base maps and other biodiversity information can be added to the
display with point and click techniques. Even some simple queries, analyses,
and graphical summaries can be performed on-the-fly. This product will be
published later this year on CD-ROM, with the software locked to the database,
making it a self-executing package that will run on standard PCs.
World Wide Web
Web browsers have become the tool of choice for many users for navigating the
Internet. Based on the hypertext model of traversing documents within a site
and across sites, browsers are easy both for users and developers. It also
has many of the built-in tools needed for our IBM project objectives to
provide better access to biodiversity datasets. Therefore, we began
development of a web home page in 1994 which can be accessed at
http://www.biogeog.ucsb.edu/. Users can select data from our archives using
two methods. The first utilizes a clickable imagemap for a region, where a
user clicks on a region on a map and receives thumbnail images of the datasets
available for that region along with a list of available datasets and
descriptions. Each dataset can be downloaded directly with the click of a
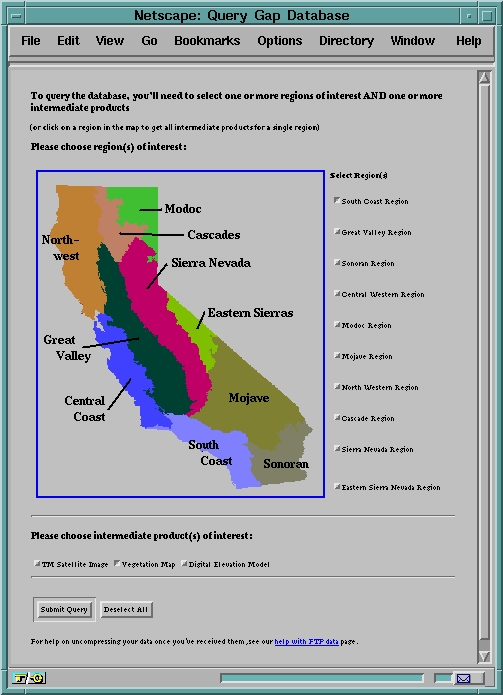
button. The second way to query the database is for the user to select both
the regions and data themes of interest by clicking checkboxes which list the
regions and themes (Figure 27). Once the datasets of interest have been
selected and the query submitted, the user sees thumbnail images of each data
theme along with a link to the real data's ftp location. The web access for
the final California GAP database will be revised shortly and data will be
available in several forms to assist users with different connection speeds or
geographic areas of interest.
Figure 27. Clickable image map on the web page for GAP database browsing
and downloading.
Next Section