Bayesian classifier of communities from species assemblages
Coastal sage scrub classification and conservation assessment
The national Gap Analysis Program is mapping vegetation alliances, which are defined by vegetation structure and dominant canopy species, using a 1 km² minimum mapping unit. Natural vegetation is rarely uniform over a square kilometer. It is more typical at this sampling scale to encounter a mosaic of two or more vegetation types in recurring stands associated with slope aspect, hillslope position, soil type and/or land use and disturbance history. For the California Gap Analysis Project, rather than generalizing this heterogeneity to a single, dominant type, we devised a strategy for encoding such areas as a vegetation mosaic in a single landscape unit. For each landscape unit, we recorded a primary vegetation cover type, which was the most widespread vegetation type or land use/land cover type in the polygon, a secondary cover type when present, a tertiary type (for some regions) and the fraction of the landscape covered by each type (10% classes). Up to 3 species were recorded for each type.
In contrast to a traditional, small-scale vegetation map, a landscape unit thus may contain information on up to three species assemblages and up to 9 plant species that are dominant or co-dominant when the vegetation is viewed over many hectares. One can query the database for distribution data on individual species, unique combinations of species, or vegetation types defined by physiognomy and/or composition. Maps of vegetation types are subsequently derived from the database by applying tables that relate each species assemblage to a particular vegetation classification system.
The vegetation database for California contains information for 21,000 map units or "landscapes." While it has been difficult to compile data on individual dominant species, and the data are uneven in their quality and timeliness, the resulting database provides an unprecedented opportunity to explore biogeographic patterns and to conduct conservation analyses for dominant plant species and species combinations. As part of our IBM-ERP research, we have tested different approaches to analyzing these species data.
We conducted an analysis of coastal sage shrublands in southwestern California to compare mapped distributional patterns of the dominant overstory species to geographic patterns of community composition that have been documented in previous, plot-based phytogeographic analyses. We also quantified the current ownership and management status of the coastal sage scrub type, its dominant species, and common species combinations. Based on a divisive information analysis of species occurrence data (Figure 19), assemblages of dominant shrub species exhibited three main distribution types, that is: south coastal, north coastal, and interior (Figure 20). However, species distribution boundaries showed very little coincidence and the pattern of species combinations was more suggestive of broad compositional gradients than of well defined types or geographic assemblages. All coastal scrub species and species complexes were mapped predominantly on private lands, many of which are under intense pressure from urban expansion. Most conservation efforts to date have focused on areas in Orange, Riverside, and San Diego Counties that are habitat for the threatened California gnatcatcher (Polioptila californica). Our analysis (Davis et al. 1994) highlighted the need to consider more northerly and interior elements as well. For example, practically all landscapes dominated by Salvia leucophylla are private lands of the western Transverse ranges, north of the current range of the gnatcatcher.
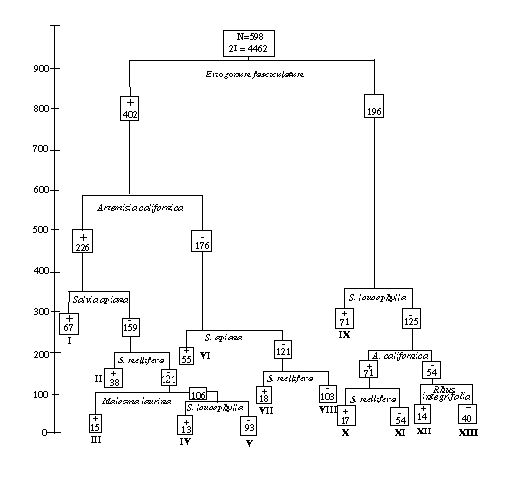
Figure 19. Divisive information classification of 598 landscapes in which coastal sage scrub is a primary or secondary vegetation type, based on species composition data. Boxes record the number of samples in each node of the tree. The height of the horizontal lines indicates the information captured by that split.
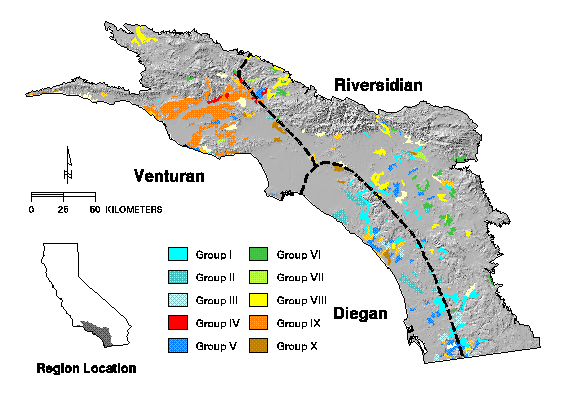
Figure 20. Composite distribution of 12 classes of coastal sage scrub identified by divisive information analysis displayed over a shaded relief image. Dashed lines are the geographic boundaries of associations as proposed by Westman (1983).
Bayesian classifier of communities from species assemblages
An inevitable problem in compiling regional vegetation databases is discrepancies among vegetation classification schemes. As described above, for the California Gap Analysis we stored preclassified data on vegetation cover and species composition in order to maintain as much flexibility as possible in relating our data to other vegetation maps. Thus we derive a vegetation class from a list of dominant species occurring in a landscape mosaic. Although simple in principle, in practice this is challenging because of the large number of unique combinations of species that we encounter at our mapping scale. We tried several algorithms to assign a particular species triplet to a class within a particular vegetation classification system.
A simple Bayesian pattern recognition approach was chosen. This technique was developed in early research on expert systems. Basically, we have characterized each vegetation class by its likelihood of containing particular species. Then when given a list of species, we can work backwards (using Bayes' rule) to determine which vegetation class is most likely to contain that particular set of species, and thus assign the class on that basis. That is, rather than trying to construct a rulebase that associates a particular species with a set of possible vegetation classes, we construct a rulebase that lists the species associated with each vegetation class.
We first used this method to translate species information in the vegetation polygons for the Sierra Nevada region into a standard community classification of vegetation in California. Based on the descriptions in this system, for each class we have rated the species most commonly found as dominants as to their relative likelihood to be present in the class. We have also given every species in the database a default rating of its likelihood to be present in classes where it is not specifically identified as a dominant. These ratings are stored in a simple ASCII table that is read as input into a Perl script which we have written to classify the species triplets.
This method has performed reasonably well for the GAP database in several regions in California. With over 1600 unique combinations of species triplets in the Sierra Nevada region, the method provided an economical and efficient way to classify each of these triplets. The method is not perfect and occasionally misclassifies triplets relative to human interpretation. But often this is because more information, such as edaphic characteristics, is needed to identify a class, or that the class has been inadequately described in the original system. After review of the probabilities of class assignments, a final set of classification assignments were made into a look-up table in a second Perl script.
Next Section